Mapping lipid C=C location isomers in organ tissues by coupling photochemical derivatization and rapid extractive mass spectrometry
Lipid desaturation plays important roles in biological processes and the disease states. Here, we report a simple but efficient method for mapping unsaturated phospholipids including the spatial distribution of lipid C=C location isomers in animal organs by coupling the C=C specific derivatization with direct analysis mass spectrometry (MS). Lipids are sampled directly by a stainless-steel wire from rat brain or kidney, extracted, and derivatized via the Paternò–Büchi reaction in a glass emitter of the nanoelectrospray ionization (nanoESI) source. Subsequent analysis by nanoESI-tandem mass spectrometry reveals C=C locations and relative quantities of lipid C=C location isomers. Unsaturated lipids, such as phospholipids and free fatty acids, have been identified with ion intensities spanning two orders of magnitude in rat brain. Typical sample consumption is less than 10 μg/measurement and the time for each analysis is about 3 min. This method should serve as a complementary method to high spatial resolution mass spectrometry imaging techniques, because it offers a streamlined experimental workflow for rapid profiling of lipids with C=C specificity to enable such applications as point-of-care disease diagnostics.
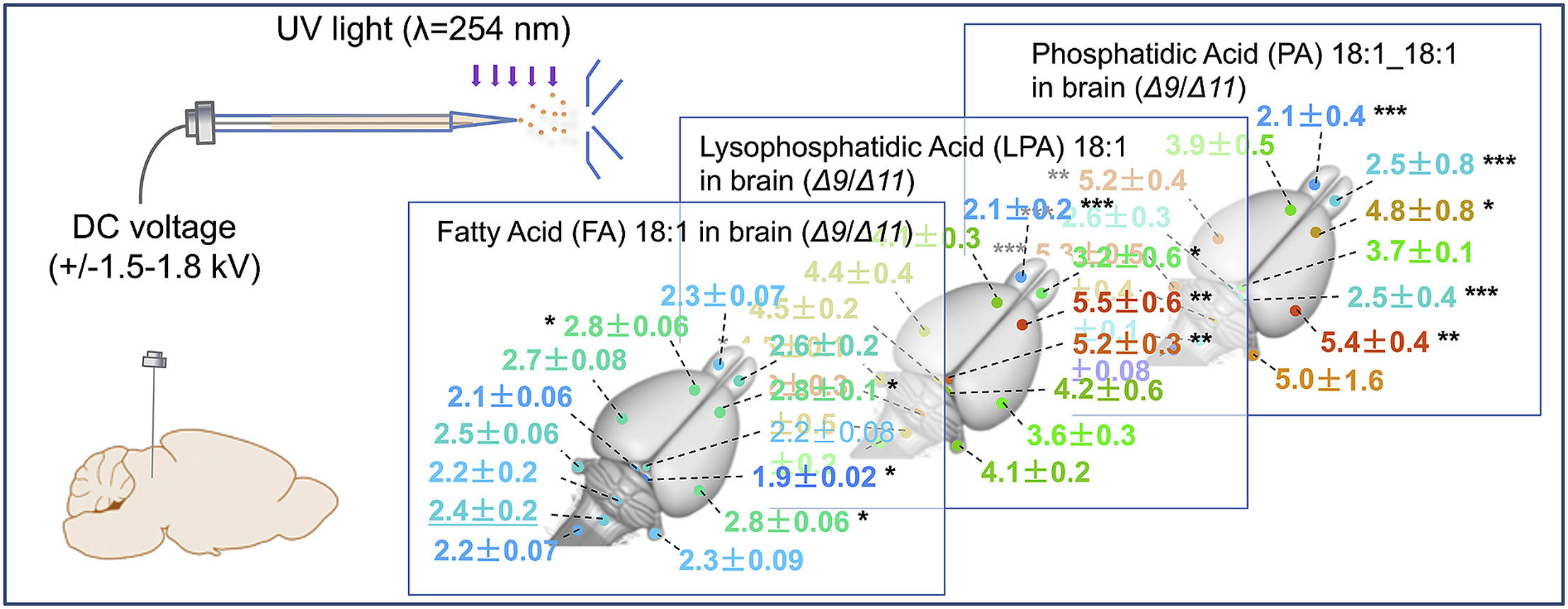
- This study establishes a streamlined workflow for rapid lipid profiling with C=C specificity in organ tissues, combining with in-situ sampling, online Paternò-Büchi derivatization and mass spectrometry analysis.
- The method enables spatial mapping of relative abundances of lipid C=C location isomers across tissue sections, with less than 10 μg sample consumption and only 3-minute analysis time.
Int J Mass Spectrom. 2019 Nov:445:116206. (IF: 1.6)


