In-Depth Characterization of Sphingoid Bases via Radical-Directed Dissociation Tandem Mass Spectrometry
Sphingoid base (SPH) is a basic structural unit of all classes of sphingolipids. A sphingoid base typically consists of an aliphatic chain that may be desaturated between C4 and C5, an amine group at C2, and a variable number of OH groups located at C1, C3, and C4. Variations in the chain length and the occurrence of chemical modifications, such as methyl branching, desaturation, and hydroxylation, lead to a large structural diversity and distinct functional properties of sphingoid bases. However, conventional tandem mass spectrometry (MS/MS) via collision-induced dissociation (CID) faces challenges in characterizing these modifications. Herein, we developed an MS/MS method based on CID-triggered radical-directed dissociation (RDD) for in-depth characterization of sphingoid bases. The method involves derivatizing the sphingoid amine with 3-(2,2,6,6-tetramethylpiperidin-1-yloxymethyl)-picolinic acid 2,5-dioxopyrrolidin-1-yl ester (TPN), followed by MS2 CID to unleash the pyridine methyl radical moiety for subsequent RDD. This MS/MS method was integrated on a reversed-phase liquid chromatography–mass spectrometry workflow and further applied for in-depth profiling of total sphingoid bases in bovine heart and Caenorhabditis elegans. Notably, we identified and relatively quantified a series of unusual sphingoid bases, including SPH id17:2 (4,13) and SPH it19:0 in C. elegans, revealing that the metabolic pathways of sphingolipids are more diverse than previously known.
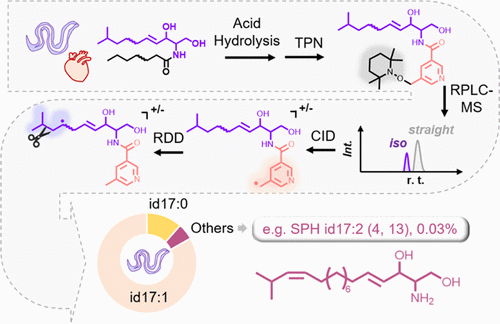
- A tandem mass spectrometry (MS/MS) method based on CID-triggered radical-directed dissociation (RDD) is developed for detailed structure elucidation of sphingoid bases.
- A reversed-phase liquid chromatography-mass spectrometry (RPLC)-MS/MS workflow is established to facilitate in-depth profiling of sphingoid bases released from lipid extracts of bovine heart and Caenorhabditis elegans.
- A series of unusual sphingoid bases, including SPH id17:2 (4,13) and SPH it19:0 in C. elegans, were identified and relatively quantified, indicating that the metabolic pathways of sphingolipids are more diverse than previously known.
J Am Soc Mass Spectrom. 2023 Oct 4;34(10):2394-2402. (IF: 3.1)


