Localization of Intrachain Modifications in Bacterial Lipids Via Radical-Directed Dissociation
Intrachain modifications of membrane glycerophospholipids (GPLs) due to formation of the carbon–carbon double bond (C═C), cyclopropane ring, and methyl branching are crucial for bacterial membrane homeostasis. Conventional collision-induced dissociation (CID) of even-electron ions of GPL favors charge-directed fragmentation channels, and thus little structurally informative fragments can be detected for locating intrachain modifications. In this study, we report a radical-directed dissociation (RDD) approach for characterization of the intrachain modifications within phosphoethanolamines (PEs), a major lipid component in bacterial membrane. In this method, a radical precursor that can produce benzyl or pyridine methyl radical upon low-energy CID at high efficiency is conjugated onto the amine group of PEs. The carbon-centered radical ions subsequently initiate RDD along the fatty acyl chain, producing fragment patterns key to the assignment and localization of intrachain modifications including C═C, cyclopropane rings, and methyl branching. Besides intrachain fragmentation, RDD on the glycerol backbone produces fatty acyl loss as radicals, allowing one to identify the fatty acyl chain composition of PE. Moreover, RDD of lyso-PEs produces radical losses for distinguishing the sn-isomers. The above RDD approach has been incorporated onto a liquid chromatography–mass spectrometry workflow and applied for the analysis of lipid extracts from Escherichia coli and Bacillus subtilis.
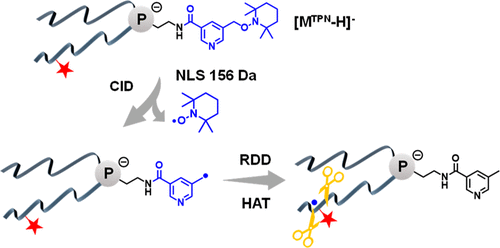
- This study develops a novel radical-directed dissociation approach triggered by low energy CID to characterize intrachain modifications (C=C, cyclopropane, methyl branching) in glycerophosphoethanolamines (PEs) and Lyso-PEs.
- This approach was applied to the analysis of Escherichia coli and PLA2-treated Bacillus subtilis lipid extracts leading to 22 identified PE species (including 6 cyclopropyl PE, 10 unsaturated PE, and 3 PE containing both cyclopropane and C=C,) and 14 LPEs with methyl branching, demonstrating utility in bacterial membrane lipidomics.
J Am Soc Mass Spectrom. 2022 Apr 6;33(4):714-721. (IF: 3.1)


